Translational Perioperative and Pain Medicine (ISSN: 2330-4871)
ARTICLE DOI: 10.31480/2330-4871/038
ORIGINAL RESEARCH OPEN ACCESS
Altered T-UCRs expression profile in the spinal cord of mice with neuropathic pain
Bao-Chun Jiang1,#, Tian Yang1,#, BS, Li-Na He1, BS,Yuan-Xiang Tao2, MD, PhD, Yong-Jing Gao, MD, PhD1,3
1Pain Research Laboratory, Institute of Nautical Medicine, Jiangsu Key Laboratory of Inflammation and Molecular Drug Target, Nantong University, Nantong, Jiangsu 226019, China
2Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA
3Co-innovation Center of Neuroregeneration, Nantong University, Nantong, Jiangsu 226001, China
#These authors contributed equally to this work.
Yong-Jing Gao, Institute of Nautical Medicine, Nantong University, 9 Seyuan Road, Nantong, Jiangsu 226019, China. Tel: +86-513-55003374; Fax: +86-513-55003370, E-mail: gaoyongjing@hotmail.com; gaoyongjing@ntu.edu.cn
Editor: Renyu Liu, MD; PhD; Associate Professor, irector of Preoperative Medicine, Department of Anesthesiology and Critical Care; erelman School of Medicine at the University of Pennsylvania, 336 John Morgan building, 3620 Hamilton Walk, Philadelphia, PA 19104, Phone: 2157461485; FAX: 2153495078, Email: liur@uphs.upenn.edu
Received: April 16, 2016 | Accepted: May 08, 2016 | Published: May 20, 2016
Citation: Jiang BC, Yang T, He LN, Tao YX, Gao YJ. Altered T-UCRs expression profile in the spinal cord of mice with neuropathic pain. Transl Perioper & Pain Med 2016; 1(3): 1-10.
Abstract
Spinal cord plays an important role in the transmission and modulation of nociceptive information. Global changes in gene expression in the spinal cord contribute to the induction and maintenance of neuropathic pain. Transcribed Ultraconserved Regions (T-UCRs), a novel class of long noncoding RNAs, can regulate gene expression at both transcriptional and post-transcriptional levels and are related to many human diseases such as cancer, Alzheimer's disease, and heart diseases. In this study, we screened abnormal T-UCRs expression in the spinal cord under spinal nerve ligation (SNL)-induced neuropathic pain condition. Microarray data showed the alternation of T-UCRs at the transcriptional level in the spinal cord 10 days after SNL. Among 78 altered T-UCRs, 23 T-UCRs were upregulated by more than 1.5-fold and 55 ones downregulated by less than 0.5-fold after SNL. Hierarchical cluster analysis of T-UCRs expression profiles showed the opposite expression pattern between SNL and sham-operated mice. The quantitative real-time reverse transcription polymerase chain reaction analysis further confirmed the expression patterns of uc.305, uc.189, uc.46, and uc.217 after SNL. The gene ontology annotation and signaling pathway analysis for the T-UCRs host genes indicated that differentially expressed T-UCRs were involved in several intracellular activities and signaling pathways, including Ephrin receptor activity, signaling pathway. Collectively, the current data suggest the possible role of T-UCR in the pathogenesis of neuropathic pain. T-UCRs may serve as a new kind of target for the treatment of neuropathic pain.
Keywords
Neuropathic pain, Spinal cord, T-UCR, Gene expression
Introduction
Neuropathic pain is a common and intractable chronic pain caused by injury or disease of the nervous system, and its pathogenesis is complex and still unclear. The synaptic and cellular mechanisms of central sensitization in the spinal cord participate in the development and maintenance of neuropathic pain [1-3]. Spinal cord central sensitization are believed to result from the differential expression of multiple pain-associated molecules and the changes of the signaling pathways after nerve injury [4]. Understanding the molecular mechanisms underlying neuropathic pain might provide novel approaches for the development of analgesic strategies.
Transcribed ultraconserved regions (T-UCRs) are a newly discovered class of regulatory non-coding RNAs. UCRs are DNA segments more than 200 bp in length, and are completely conserved among human, rat, and mouse [5, 6]. The 100% conservation of T-UCRs among mammalian genomes indicates that T-UCRs are functionally important in the regulation of gene expression. Indeed, previous studies have shown that hundreds of T-UCRs in the genome of human, mouse and rat are involved in the regulation of gene expression at both transcriptional and posttranscriptional levels [7]. T-UCRs that overlapped with coding exons are classified as exonic type, and the remaining ones as non-exonic T-UCRs. Exonic UCRs play an important role in post-transcriptional regulation, such as alternative splicing and mRNA processing [7]. Non-exonic T-UCRs typically occur near genes encoding transcription factors, and control the expression of adjacent transcription factors at both the DNA and RNA levels [8, 9]. Most recent detective techniques and genome-wide microarray profiling have shown the transcribed UCRs with distinct signatures during development [10, 11] as well as under some disease conditions [12]. However, the genome-wide expression and functional significance of T-UCRs in neuropathic pain remain unclear.
In the present study, we compared the expression pattern of T-UCRs in the spinal cord between L5 spinal nerve ligation (SNL)-treated and sham surgery-treated mice using microarray method. We identified 78 differentially expressed (DE) T-UCRs. Furthermore, gene ontology (GO) and signaling pathway analyses of the DE T-UCRs' overlapping genes indicate that these T-UCRs may play a functional role in neuropathic pain.
Materials and Methods
Animals and surgery
Adult male ICR mice (male, 8 weeks) were purchased from Experimental Animal Center of Nantong University. The animals were maintained on a 12:12 light–dark cycle at a room temperature of 22 ± 1℃ with free access to food and water. The experimental procedures were approved by the Animal Care and Use Committee of Nantong University and performed in accordance with the guidelines of the International Association for the Study of Pain. For the SNL model, animals were anesthetized with isoflurane and the L6 transverse process was removed to expose the L4 and L5 spinal nerves. The L5 spinal nerve was then isolated and tightly ligated with 6-0 silk thread [13]. For sham operations, the L5 spinal nerve was exposed but not ligated.
RNA extraction
Total RNA was extracted from the ipsilateral L5 spinal cord 10 days post-surgery using the Trizol method (Invitrogen, Life Technologies). Three ipsilateral mouse spinal cords were pooled together to achieve enough RNA. Totally 18 mice were used for RNA extraction (9 for SNL and 9 for sham). Following extraction, RNA was further purified with a NucleoSpin® RNA clean-up kit (MACHEREY-NAGEL, Germany). The concentrations and quanlities of the RNA samples were determined by a NanoDrop ND-2000 Spectrophotometer (Thermo Scientific, Waltham, MA). The ratio of 28S:18S band intensities were detected by formaldehyde denaturalization electrophoresis. The samples with total RNA quality > 5 μg or the 28S:18S ratio > 2 were further examined.
Microarray of T-UCRs
T-UCRs expression profiles in the spinal cord were assessed using Arraystar Mouse LncRNA Microarray V2.0 (8×60 K, Arraystar), which can profile the global expression of mice T-UCRs. For each sample, T7 Oligo (dT) primer containing T7 RNA polymerase promoter sequence was used to synthesize the first strand cDNA from total RNA by CbcScript reverse transcriptase. The RNase H-DNA polymeraseⅠ controlled the process of the second-strand cDNA synthesis (KangChen Bio-tech, China). T7 promoter drove the synthesis of cRNA using the above cDNA as the template. The cRNA were reverse-transcribed into cDNA again by random primers, and then the transcription products were labeled in the presence of Cy3-dCTP, random primers, and Klenow Enzyme. The microarray slides were hybridized at 45℃ for 16 h in 2 × GEx Hybridization Buffer HI-RPM (Agilent Technologies) containing the labeled cDNA. After being washed, microarray results were scanned by Agilent G2505B Microarray Scanner, and images were quantified using Agilent's Feature Extraction software (Agilent Technologies). Quantile normalization of raw data and subsequent data processing was performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies).
Analysis of microarray data
To determine the significant differentially expressed T-UCRs after SNL, raw intensity values were normalized and analyzed using the Significance Analysis of Microarrays software (SAM, version 2.1, Stanford University, CA). Significance was assigned to the T-UCRs which ratio of SNL to sham is more than 1.5 or less than 0.5. Hierarchical cluster analysis was carried out for all T-UCRs or differentially expressed T-UCRs using cluster 3.0 and visualized in Java Treeview. To search the potential functions of differentially expressed T-UCRs in neuropathic pain, their host genes were subjected to Molecule Annotation System CapitalBio MAS 3.0 software (CapitalBio Corporation, Beijing, China) to perform signaling pathways and GO molecular function terms analysis.
Quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) analysis of T-UCRs
To validate the microarray data, quantitative real-time RT-PCR assay was used to examine the expression of 4 T-UCRs including uc305, uc.189, uc.46, and uc.217. Total RNA from the spinal cord was extracted as described above. One microgram of total RNA was reverse-transcribed using random 6 mer primers according to the manufacturer's protocol (Takara, Shiga, Japan). PCR analysis was conducted in the Real-time Detection System (Rotor-Gene 6000, Hamburg, Germany) by SYBR green I dye detection (Takara). The primer sequences for each gene are shown in Table 1. The PCR amplifications were performed at 95 ℃ for 30 s, followed by 40 cycles of thermal cycling at 95 ℃ for 5 s and 60 ℃ for 45 s. GAPDH RNA were used as endogenous control to normalize differences for mRNA detection. Melt curves were performed on completion of the cycles to ensure that nonspecific products were absent. Quantification was done by normalizing Ct (cycle threshold) values with GAPDH Ct and analyzed with the 2-ΔΔCT method.
Table 1: Primer sequences and information used in Real-Time PCR.
| UCR Name | Sequence (5'~3') | Amplicon size |
|---|---|---|
| uc.305 | Forward:CAGCCTCACCTCACCATTTCT Reverse:AGCAGTAAACTGTATAAGCAGGA |
71 bp |
| uc.189 | Forward:AGTCGACTGTTTTCTTTAGTAGGCT Reverse:AAAAAGGAACCCGCAGGCA |
75 bp |
| uc.46 | Forward:CCTGTAAAATCTACGGAGCGGA Reverse:AGAGTACAGGGGTGCAGAATG |
72 bp |
| uc.217 | Forward:CAGCAGACACCAGCGAATAA Reverse:TTGACATTATCATTAAGAGTGGGTG |
72 bp |
Statistical analysis
The data are expressed as means±SEM. Differences between the groups were compared by Student's t-test. The criterion for statistical significance was P < 0.05.
Results
In the present study, we used SNL-induced neuropathic pain model to investigate the expression profiling of T-UCRs. As reported previously, SNL produced remarkable pain hypersensitivities including mechanical allodynia and heat hyperalgesia, with robust glial activation in the ipsilateral dorsal horn of the spinal cord on day 10 post-SNL [14]. We harvested the ipsilateral spinal cord 10 days after SNL or sham surgery and then carried out the microarray analysis. This standard array with 606 probe sets could detect the expression of all 473 T-UCRs genes including both exonic and nonexonic forms. We found that majority of the transcribed UCRs were expressed in the spinal cord of either sham or SNL mice. In order to identify putative T-UCRs involved in neuropathic pain, we compared the data with student's t-test and set P value as < 0.05 and fold change as > 1.5 or < 0.5. We found that 23 T-UCRs were upregulated and 55 ones down-regulated after SNL. These T-UCRs were further defined by their relative positions to neighbored protein-coding gene. Foutytwo T-UCRs (54%) were sense overlapping T-UCRs, 20 (26%) intergenic T-UCRs, and 16 (21%) antisense overlapping T-UCRs. While ranking the normalized signal intensities for expression levels, we identified that uc.35, uc.99, uc.211, uc.153, uc.257, uc.1, uc.157, uc.137, and uc.107 were mostly up-regulated and that uc.300, uc.158, uc.184, uc.217, uc.252, uc.33, uc.264, uc.50, uc.197, and uc.46 were mostly down-regulated (Table 2). These results indicate that nerve injury led to the distinct change in T-UCRs' expression in the ipsilateral spinal cord.
Table 2: Most significant differentially expressed T-UCRs in the spinal cord.
| UCR name | Probe name | Regulation | Type | Location | Significance | Neighboring gene |
|---|---|---|---|---|---|---|
| uc.99 | CUST_97_PI426073487 | up | intergenic | chr2 | 0.000420468 | Dlx1 |
| uc.211 | CUST_690_PI426073487 | up | sense overlap | chr6 | 0.0805771 | Skap2 |
| uc.189 | CUST_187_PI426073487 | up | sense overlap | chr17 | 0.000449672 | Srsf3 |
| uc.153 | CUST_632_PI426073487 | up | antisense overlap | chr13 | 0.000940733 | Tnpo1 |
| uc.99 | CUST_736_PI426073487 | up | antisense Overlap | chr4 | 0.000897086 | Zcchc7 |
| uc.1 | CUST_482_PI426073487 | up | antisense overlap | chr4 | 0.0068662 | Pex14 |
| uc.107 | CUST_586_PI426073487 | up | intergenic | chr2 | 0.001610238 | Sp9, Cir1 |
| uc.214 | CUST_212_PI426073487 | up | intergenic | chr6 | 0.001404882 | Neurod6 |
| uc.305 | CUST_303_PI426073487 | up | sense overlap | chr19 | 0.000917825 | Btrc |
| uc.329 | CUST_327_PI426073487 | up | antisenseoverlap | chr2 | 0.011064756 | 0610012H03Rik |
| uc.36 | CUST_34_PI426073487 | down | antisense overlap | chr3 | 0.000648519 | Prpf38b |
| uc.186 | CUST_665_PI426073487 | down | sense overlap | chr11 | 0.000219314 | Hnrnph1 |
| uc.46 | CUST_525_PI426073487 | down | sense overlap | chr1 | 0.000199143 | Hnrnpu |
| uc.197 | CUST_195_PI426073487 | down | intergenic | chr4 | 0.001758473 | |
| uc.50 | CUST_48_PI426073487 | down | antisense overlap | chr17 | 0.248408997 | Srsf7 |
| uc.264 | CUST_743_PI426073487 | down | sense overlap | chr13 | 0.001940631 | Hnrnpk |
| uc.33 | CUST_31_PI426073487 | down | sense overlap | chr3 | 6.55023E-05 | Ptbp2 |
| uc.217 | CUST_215_PI426073487 | down | sense overlap | chr11 | 0.000100225 | Vstm2a |
| uc.184 | CUST_182_PI426073487 | down | intergenic | chr11 | 0.000130604 | Nsg2 |
| uc.158 | CUST_637_PI426073487 | down | intergenic | chr13 | 0.006124622 | uc008xvd.1 |
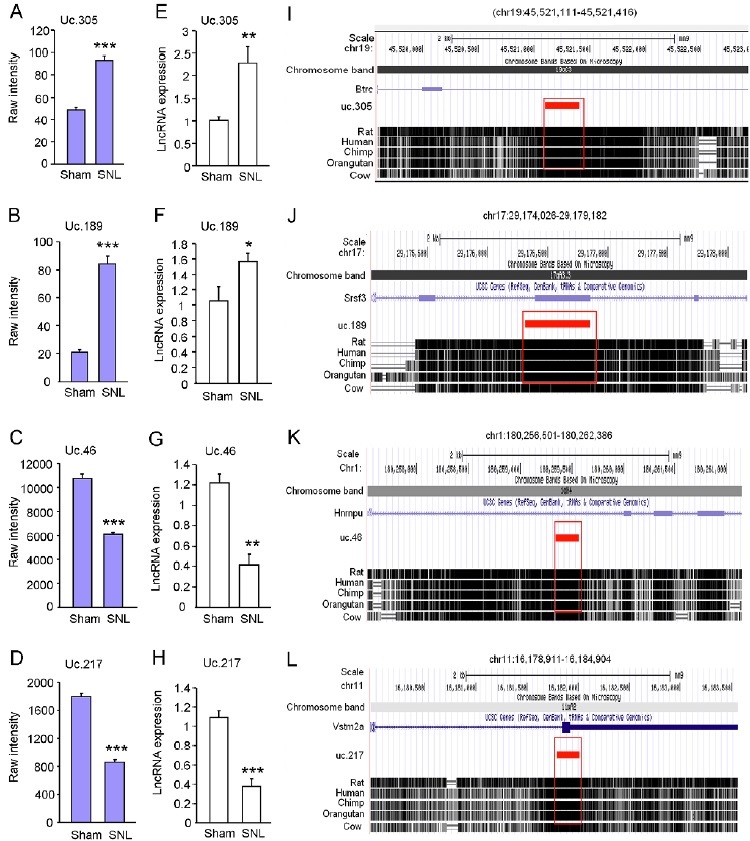
To validate the changes of T-UCRs identified by the microarray analysis, we carried out quantitative real-time RT-PCR to examine the expression of some T-UCRs in the lumbar spinal cord homogenates from another group of mice 10 days after SNL or sham-operation. Two up-regulated (uc.305 and uc.189, Fig 1. A, B) and 2 down-regulated (uc.46 and uc.217, Fig. 1 C, D) T-UCRs were chosen for the PCR detection, as the sequences of these T-UCRs are suitable for the design of the primer pairs. Consistent with the microarray data, RT-PCR analysis showed that uc.305 (Fig. 1E) and uc.189 (Fig 1F) were significantly upregulated and that uc.46 (Fig 1G) and uc.217 (Fig 1H) were markedly downregulated after SNL. Genomic locations of the above T-UCR genes indicate that they can be transcribed in either exonic or nonexonic forms to their host genes. Based on the searching results from Multiz Alignment & Conservation tracks of 5 mammal species. uc.305 (Fig. 1I), uc.189 (Fig. 1J), uc.46 (Fig. 1K), and uc.217 (Fig. 1L) each displayed 100% homology between mouse, rats, humans, chimp and orangutan.
Figure 1: Validation of microarray data by quantitative RT-PCR. (A-H) Microarray data (A-D) and RT-PCR data (E-H) show the expression of uc.305 (A, E), uc.189 (B, F), uc.46 (C, G), and uc.217 (D, H) in the spinal cord of SNL or sham-operated mice. n = 3 mice for microarray data. n = 4-5 mice for RT-PCR. (I-L) The genome location and overlapping genes for uc.305 (I), uc.189 (J), uc.46 (K), and uc.217 (L), respectively. Species conservation alignments of uc.305, uc.189, uc.46, and uc.217 DNA segments among rat, human, chimp, orangutan, and cow were identified by UCSC Genome browser. All the four T-UCRs show 100% conservation across species (mouse, rat, and human). Conserved sequences are highlighted in red rectangle.
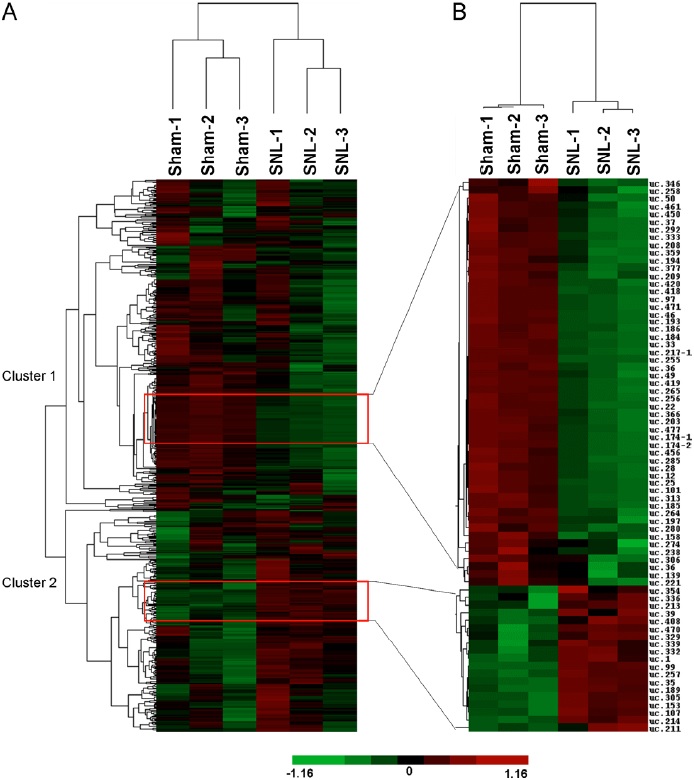
Global patterns of T-UCRs expression in the spinal cord from the SNL and sham groups were dissected using hierarchical cluster analysis and visualized as heatmaps (Fig. 2). Hierarchical cluster analysis of all T-UCRs expression profiles showed that the 3 sham samples and 3 SNL ones were clustered together, respectively, and that the signal intensities had good intraclass consistency, indicating the high repeatability of microarray data. Furthermore, T-UCRs expression could be divided into two clusters after SNL: the decreased T-UCRs (cluster 1) and the T-UCRs-transcriptome in the spinal cord. The heatmap of T-UCRs with expression changes of more than 1.5 fold or less than 0.5-fold was shown in Fig. 2B. High degree of concordance was observed in either SNL or sham samples. The number of the down-regulated T-UCRs was higher than that of the up-regulated T-UCRs.
Figure 2: Analysis of T-UCRs expression from SNL and sham mice. (A) Hierarchical clustering shows overall T-UCRs expression pattern of reliably measured probe sets (608). Rows represent individual probe sets. Columns represent individual samples. The scale bar at the bottom of the matrix indicates the degree of T-UCRs expression levels. Green indicates down-regulation. Red indicates up-regulation. Black indicates no change. The expression of T-UCRs was decreased in cluster 1 and increased in cluster 2 after SNL. (B) Heat map shows the T-UCRs whose expression is more than 1.5-fold or less than 0.5-fold after SNL.
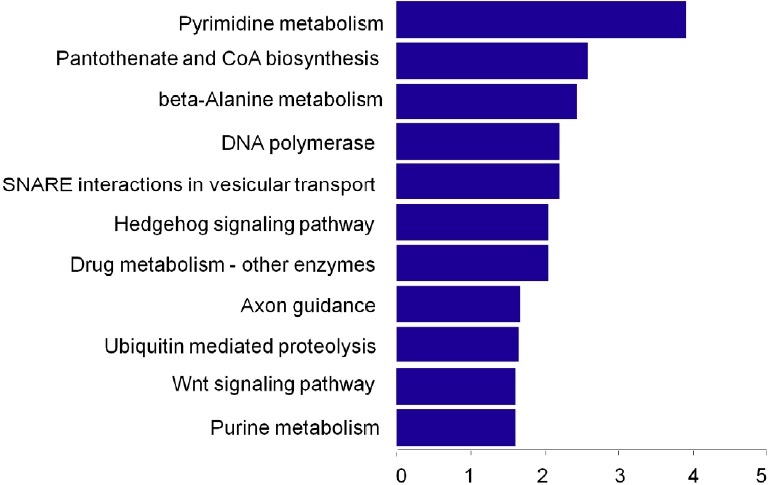
To explore the potential function of the identified T-UCRs, we conducted GO term and signaling pathway annotation analysis using Molecule Annotation System 3.0 (MAS3.0, http://bioinfo. capitalbio.com/mas/). GO term analysis highlighted the most relevant GO terms for T-UCRs-related genes including the RNA binding, chemorepellant activity, transcription factor activity, nucleotide binding, and ephrin receptor activity (Fig. 3). It indicates that the differentially expressed T-UCRs may control the expression of some genes. Signaling pathway analysis showed that these target genes were associated with the WNT signaling pathway, axon guidance, and pyrimidine metabolism pathways (Fig. 4).
Figure 3: Function prediction of 78 differentially expressed T-UCRs-related mRNAs. Significant molecular functions (A), biological process (B), and cellular component (C), respectively, were listed according to the enrichment scores (-log10 (P-value)) from high to low significance.
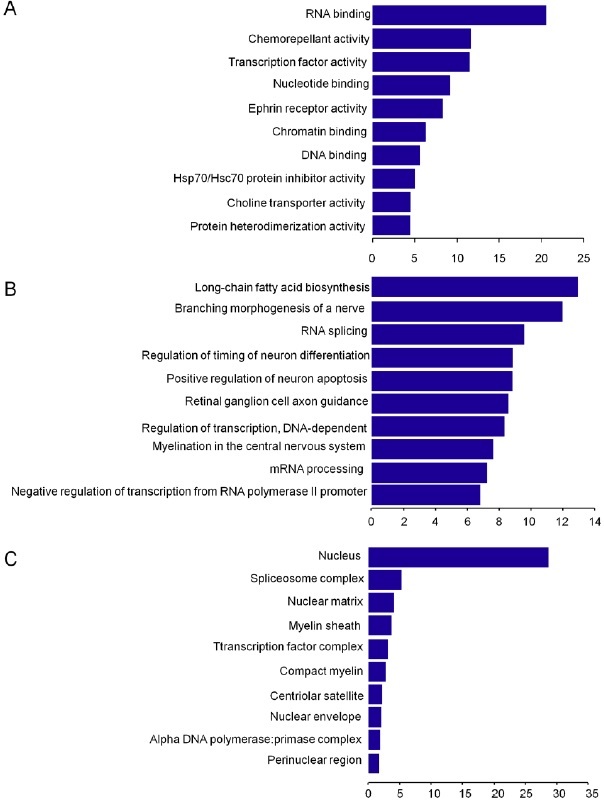
Figure 4: Pathway analysis of 78 differentially expressed T-UCRs-related mRNAs. Enrichment scores (-log10 (P-value)) of the signaling pathways were shown in histogram and listed from high to low significance.
Discussion
Previous microarray analysis has indicated dramatic changes in the expression of the genes in the painrelated regions of nervous system under neuropathic pain and inflammatory pain conditions [4,15]. Noncoding RNAs, including microRNAs [16-18] and long non-coding RNAs (lncRNA) [14, 19, 20], were upregulated or downregulated in the DRG and spinal cord. Recent studies showed that lncRNAs play a wide variety of regulatory roles in gene expression [21]. Unlike small RNAs, such as microRNAs or snoRNAs, lncRNAs identified previously are generally lack of strong conservation. This may limit their applications in clinic, especially in pain management, given that the majority of previous studies were carried out in rodent animals. T-UCRs are new class of lncRNAs. As T-UCRs are of 100% homology among human, mouse and rat, they are likely important for biomedical research [22].
Neuropathic pain is defined as chronic pain resulting from the injury or the diseases in peripheral and/ or central nervous sytem [23]. The spinal cord is one of key regions in mediating the pathogenesis of this disorder [24-26]. To our knowledge, whether the T-UCRs in the spinal cord were involved in neuropathic pain is still elusive. The high-density microarray technology allows to systematically determine the expression levels of several thousand messenger RNA or non-coding RNA transcripts concurrently. In the present study, we found that the expression of T-UCRs was significantly changed in the spinal cords of mice following SNL. These changes suggest that T-UCRs may be involved in neuropathic pain processing.
Our microarray analysis showed that 23 T-UCRs were up-regulated and 55 ones down-regulated in the spinal cord on day 10 after SNL. uc.99, one of the most upregulated T-UCRs, may regulate the expression of distal-less homeobox 1 (Dlx1). The latter encodes one of a homeobox transcription factor gene family and control the neuron-glial switch in the developing forebrain [27]. The level of uc.305 was increased by about 2 folds after SNL. As a host gene of uc. 305, Btrc (beta-transducin repeat containing E3 ubiquitin protein ligase) is a member of WNT signaling pathway. Loss of Btrc function results in the stabilization of β-catenin and the activation of WNT signaling [28]. Spinal blockade of WNT signaling pathways inhibited the production and persistence of neuropathic pain and the accompanying neurochemical alterations without affecting normal pain sensitivity and locomotor activity [29]. It is very likely that uc.305 may be involved in neuropathic pain by regulating the WNT signaling pathway.
In order to fully understand the function of the differentially expressed T-UCRs, GO functional enrichment and signaling pathway annotation were applied to their target gene pool. GO term annotation results showed that RNA binding, chemorepellant activity, transcription factor activity, nucleotide binding and ephrin receptor activity were the most significantly enriched GO terms. Interestingly, T-UCRassociated genes exhibit nucleotide binding, chromatin binding, DNA binding, and transcription factor activity, indicating that the T-UCRs might be involved in epigenetic and transcriptional regulation during pain processing. Pathway annotation showed a significant association of T-UCRs with the axon guidance, pyrimidine metabolism, and WNT signaling pathway in neuropathic pain, indicating that differentially expressed T-UCRs may participate in pain modulation by regulating protein-coding genes in these pathways.
Conclusion
Our microarray results demonstrated for the first time that the expression profile of T-UCRs in the spinal cord was altered in SNL-induced neuropathic pain. These aberrantly expressed T-UCRs may participate in the induction and maintenance of neuropathic pain. Further studies are required to determine whether these T-UCRs can serve as novel therapeutic targets or diagnostic biomarkers for this disorder.
Disclosure of Funding
This study was supported by grants from the National Natural Science Foundation of China (31371121 and 81400915 ) to Y.J.G and B.C.J., from the Natural Science Foundation of Jiangsu Province (BK20140427) to B.C.J., and by NIH grants (R01NS094664, R01DA033390, and U01HL117684) to Y.X.T.
Conflict Interests Disclosure
The authors have no conflicting interests to disclose.
References
- Kawasaki Y, Zhang L, Cheng J-K, Ji R-R. Cytokine mechanisms of central sensitization: distinct and overlapping role of interleukin-1β, interleukin-6, and tumor necrosis factor-α in regulating synaptic and neuronal activity in the superficial spinal cord. The Journal of neuroscience. 2008;28(20):5189-94.
- Woolf CJ, Thompson SW. The induction and maintenance of central sensitization is dependent on N-methyl-D-aspartic acid receptor activation; implications for the treatment of post-injury pain hypersensitivity states. Pain. 1991;44(3):293-9.
- Gao Y-J, Zhang L, Samad OA, Suter MR, Yasuhiko K, Xu Z-Z, et al. JNK-induced MCP-1 production in spinal cord astrocytes contributes to central sensitization and neuropathic pain. The Journal of neuroscience. 2009;29(13):4096-108.
- LaCroix-Fralish ML, Austin JS, Zheng FY, Levitin DJ, Mogil JS. Patterns of pain: meta-analysis of microarray studies of pain. Pain. 2011;152(8):1888-98.
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved elements in the human genome. Science. 2004;304(5675):1321-5.
- Harmston N, Barešić A, Lenhard B. The mystery of extreme non-coding conservation. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368(1632):20130021.
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & development. 2007;21(6):708-18.
- Sandelin A, Bailey P, Bruce S, Engström PG, Klos JM, Wasserman WW, et al. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC genomics. 2004;5(1):99.
- Licastro D, Gennarino VA, Petrera F, Sanges R, Banfi S, Stupka E. Promiscuity of enhancer, coding and non-coding transcription functions in ultraconserved elements. BMC genomics. 2010;11(1):151.
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes & development. 2006;20(11):1470-84.
- Sanges R, Hadzhiev Y, Gueroult-Bellone M, Roure A, Ferg M, Meola N, et al. Highly conserved elements discovered in vertebrates are present in nonsyntenic loci of tunicates, act as enhancers and can be transcribed during development. Nucleic Acids Research. 2013;41(6):3600-18.
- Peng JC, Shen J, Ran ZH. Transcribed ultraconserved region in human cancers. RNA biology. 2013;10(12):1771-7.
- Ho Kim S, Mo Chung J. An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain. 1992;50(3):355-63.
- Jiang BC, Sun WX, He LN, Cao DL, Zhang ZJ, Gao YJ. Identification of lncRNA expression profile in the spinal cord of mice following spinal nerve ligation-induced neuropathic pain. Molecular pain. 2015;11(1):43.
- Lutz BM, Bekker A, Tao YX. Noncoding RNAs: new players in chronic pain. Anesthesiology. 2014;121(2):409-17.
- Park CK, Xu ZZ, Berta T, Han Q, Chen G, Liu XJ, et al. Extracellular MicroRNAs Activate Nociceptor Neurons to Elicit Pain via TLR7 and TRPA1. Neuron. 2014;82(1):47-54.
- Brandenburger T, Castoldi M, Brendel M, Grievink H, Schlösser L, Werdehausen R, et al. Expression of spinal cord microRNAs in a rat model of chronic neuropathic pain. Neuroscience letters. 2012;506(2):281-6.
- Sakai A, Saitow F, Miyake N, Miyake K, Shimada T, Suzuki H. miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain. 2013;136(9):2738-50.
- Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao J-Y, Liang L, et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nature neuroscience. 2013;16(8):1024-31.
- Wu S, Marie Lutz B, Miao X, Liang L, Mo K, Chang YJ, et al. Dorsal root ganglion transcriptome analysis following peripheral nerve injury in mice. Molecular pain. 2016;12.
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature structural & molecular biology. 2013;20(3):300-7.
- Mestdagh P, Fredlund E, Pattyn F, Rihani A, Van Maerken T, Vermeulen J, et al. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene. 2010;29(24):3583-92.
- Treede R-D, Jensen TS, Campbell J, Cruccu G, Dostrovsky J, Griffin J, et al. Neuropathic pain redefinition and a grading system for clinical and research purposes. Neurology. 2008;70(18):1630-5.
- Bradesi S. Role of spinal cord glia in the central processing of peripheral pain perception. Neurogastroenterology & Motility. 2010;22(5):499-511.
- Boroujerdi A, Zeng J, Sharp K, Kim D, Steward O, Luo ZD. Calcium channel alpha-2-delta-1 protein upregulation in dorsal spinal cord mediates spinal cord injury-induced neuropathic pain states. Pain. 2011;152(3):649-55.
- Nielsen LA, Henriksson KG. Pathophysiological mechanisms in chronic musculoskeletal pain (fibromyalgia): the role of central and peripheral sensitization and pain disinhibition. Best practice & research Clinical rheumatology. 2007;21(3):465-80.
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. Dlx1 and Dlx2 Control Neuronal versus Oligodendroglial Cell Fate Acquisition in the Developing Forebrain. Neuron. 2007;55(3):417-33.
- Wolter M, Scharwächter C, Reifenberger J, Koch A, Pietsch T, Reifenberger G. Absence of detectable alterations in the putative tumor suppressor gene BTRC in cerebellar medulloblastomas and cutaneous basal cell carcinomas. Acta neuropathologica. 2003;106(4):287-90.
- Zhang YK, Huang ZJ, Liu S, Liu YP, Song AA, Song XJ. WNT signaling underlies the pathogenesis of neuropathic pain in rodents. J Clin Invest. 2013;123(5):2268-86.